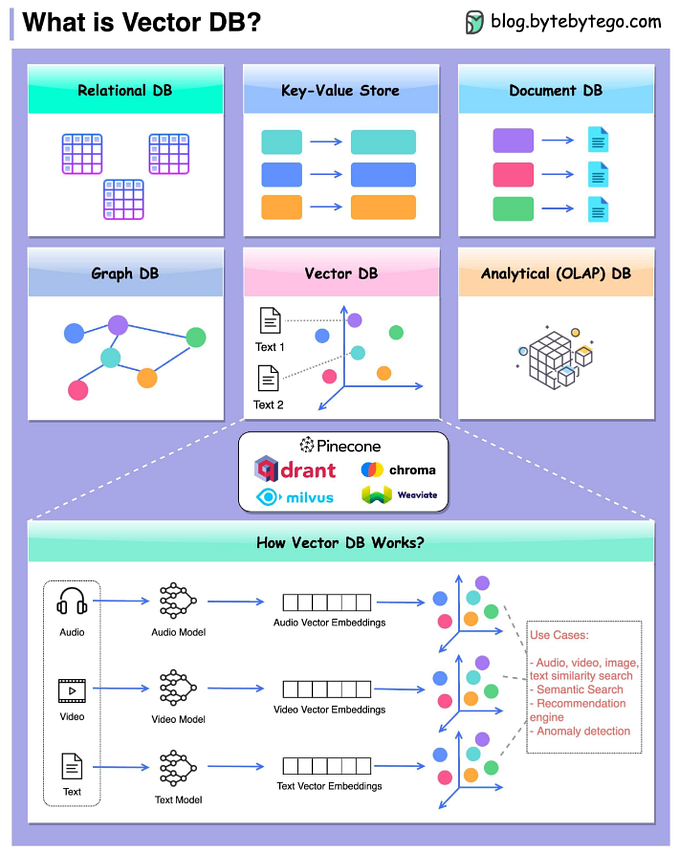
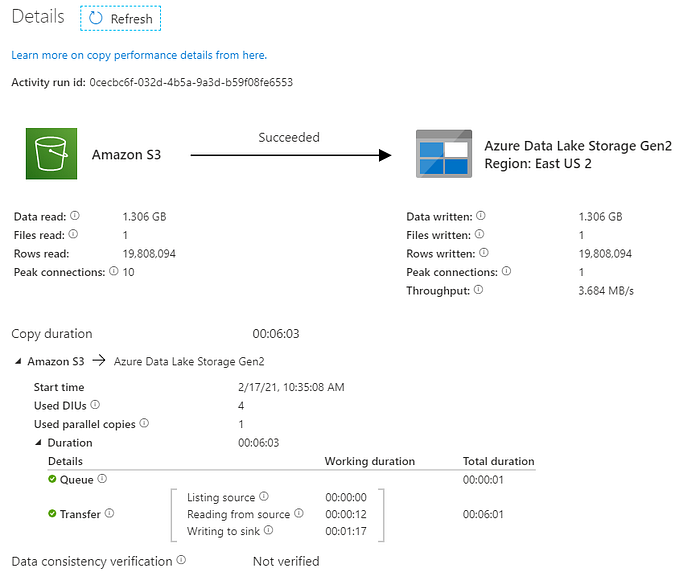
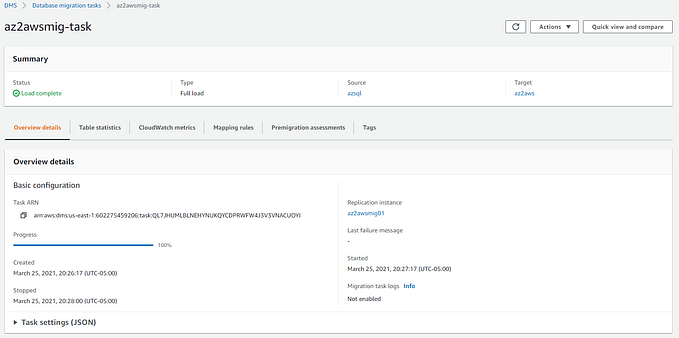
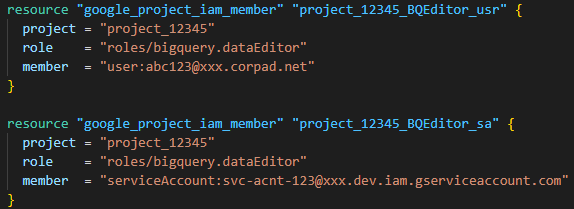
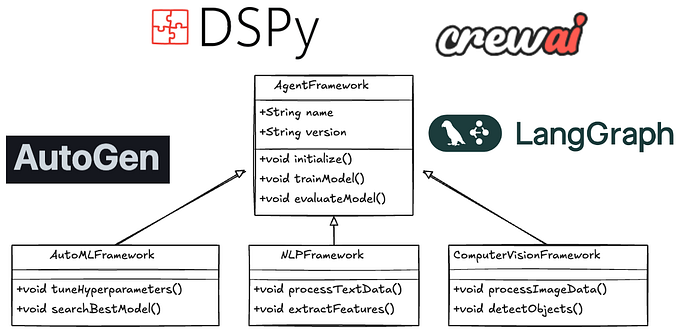
Multi-model Clinical AI agent
At their core, AI agents are systems designed to perform tasks, make decisions, and interact with their environment autonomously, often mimicking human-like behavior.
The buzz around AI agents is at an all-time high, and it’s not hard to see why. Recent breakthroughs in artificial intelligence — powered by innovations in deep learning, reinforcement learning, and natural language processing — have supercharged their capabilities. Today’s AI agents are more than just tools; they are collaborators, capable of understanding context, solving complex problems, and anticipating user needs.
In this blog post, I’ll share details about how I developed a simple multi-model clinical AI agent. This is my tech stack.
- Azure OpenAI and Gemini 2.0 Flash Experimental
- CrewAI
- MIMIC-III Clinical database
Enabling the LLMs for the PoC
- Azure OpenAI: By integrating OpenAI’s cutting-edge capabilities with Azure’s enterprise-grade infrastructure, Azure OpenAI service enables businesses to build intelligent applications with enhanced scalability, security, and compliance.
- Google Gemini: A GCP project is needed to use Gemini. This link provides details on how to install and use Gemini LLM.

Getting data from the MIMIC-III Clinical database
MIMIC-III is an extensive, freely available database comprising de-identified health-related data associated with over forty thousand patients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. More details about the database can be found here.
I ran this SQL statement to get the clinical notes of 100 patients. I exported the data to a CSV file and used it in my code, as PhysioNet doesn’t allow you to run BigQuery jobs directly on the MIMIC-III Clinical database.
WITH
patient_subset AS (
SELECT
DISTINCT subject_id
FROM
`physionet-data.mimiciii_clinical.patients`
ORDER BY
subject_id
LIMIT
100 ),
bnp_data AS (
SELECT
ce.subject_id,
ce.hadm_id,
ce.charttime AS bnp_time,
ce.valuenum AS bnp_level
FROM
`physionet-data.mimiciii_clinical.chartevents` ce
JOIN
`physionet-data.mimiciii_clinical.d_items` di
ON
ce.itemid = di.itemid
WHERE
LOWER(di.label) LIKE '%bnp%'
AND ce.valuenum IS NOT NULL ),
combined_notes AS (
SELECT
ne.subject_id,
ne.hadm_id,
STRING_AGG(ne.text, ' ') AS all_clinical_notes
FROM
`physionet-data.mimiciii_notes.noteevents` ne
JOIN
patient_subset ps
ON
ne.subject_id = ps.subject_id
GROUP BY
ne.subject_id,
ne.hadm_id )
SELECT
p.subject_id,
p.gender,
p.dob,
a.hadm_id,
a.admittime,
a.dischtime,
a.hospital_expire_flag,
dc.drg_code,
dc.description AS drg_description,
d.icd9_code AS diagnoses_icd9_codes,
bn.bnp_level,
bn.bnp_time,
cn.all_clinical_notes
FROM
patient_subset ps
JOIN
`physionet-data.mimiciii_clinical.patients` p
ON
ps.subject_id = p.subject_id
JOIN
`physionet-data.mimiciii_clinical.admissions` a
ON
p.subject_id = a.subject_id
LEFT JOIN
`physionet-data.mimiciii_clinical.drgcodes` dc
ON
a.hadm_id = dc.hadm_id
LEFT JOIN
`physionet-data.mimiciii_clinical.diagnoses_icd` d
ON
a.hadm_id = d.hadm_id
LEFT JOIN
bnp_data bn
ON
a.hadm_id = bn.hadm_id
LEFT JOIN
combined_notes cn
ON
a.hadm_id = cn.hadm_id;Agents, Tasks and Crew
This course provides a good introduction to CrewAI. I developed this code to run the multi-model Agents and Tasks.
from crewai import Agent, Task, Crew, LLM, Process
from crewai_tools import ScrapeWebsiteTool
import os
from langchain import PromptTemplate
import pandas as pd
import google.auth
from google.api_core.exceptions import BadRequest
from google.cloud import aiplatform, bigquery
# Azure OpenAI setup
os.environ["AZURE_API_KEY"] = <get the key from Azure OpenAI instance>
os.environ["AZURE_API_BASE"] = <get the end point URL from Azure OpenAI instance
os.environ["AZURE_API_VERSION"] = <get the API version from Azure OpenAI instance>
# Google Gemini setup
_REGION = <appropriate GCP region name>
_CREDENTIALS, _PROJECT_ID = google.auth.default()
_JSON_HEADERS = {"Content-Type": "application/fhir+json;charset=utf-8"}
_MODEL_ID = "gemini-2.0-flash-exp"
_MODEL_MED_PALM2 = 'medlm-large'
# Load the MIMIC-III data that's exported to a csv file
file_path = "C:/desktop/mimic_drg.csv"
clinical_notes = pd.read_csv(file_path, low_memory=False)
# Define an LLM instance for Gemini
gemini_llm = LLM(model= _MODEL_ID, temperature = 0.5)
# Define an agent for Gemini
identify_drg_gemini = Agent(
role="Medical Diagnosis Specialist",
goal="Determine the correct Diagnosis-Related Group (DRG) for a patient based on the provided medical documents, "
"clinical data, diagnoses, procedures, and other relevant indicators.",
verbose=True,
memory=True,
backstory=(
"""As a medical diagnosis specialist, you have extensive experience in analyzing clinical data, notes, lab results,
procedures, and imaging studies to identify and validate Diagnosis-Related Groups (DRGs).
Your expertise ensures accurate grouping of patient cases into appropriate DRGs, detecting any discrepancies
or misclassifications in the documented data. You use advanced reasoning to correlate clinical markers with DRG criteria."""
),
llm=gemini_llm
allow_delegation=False
)
# Define an agent for Azure OpenAI
identify_drg_azure = Agent(
role="Medical Diagnosis Specialist",
goal="Determine the correct Diagnosis-Related Group (DRG) for a patient based on the provided medical documents, "
"clinical data, diagnoses, procedures, and other relevant indicators.",
verbose=True,
memory=True,
backstory=(
"""As a medical diagnosis specialist, you have extensive experience in analyzing clinical data, notes, lab results,
procedures, and imaging studies to identify and validate Diagnosis-Related Groups (DRGs).
Your expertise ensures accurate grouping of patient cases into appropriate DRGs, detecting any discrepancies
or misclassifications in the documented data. You use advanced reasoning to correlate clinical markers with DRG criteria."""
),
llm="azure/gpt-4o", # LLM instance is not created for Azure OpenAI
allow_delegation=False
)
# Define a task for Gemini
task_identify_drg_gemini = Task(
description=(
f"Analyze the medical documents of the given patient to validate the Diagnosis-Related Group (DRG) assigned. Medical documents: {clinical_notes}. "
"Use tools to analyze the patient's clinical data, including lab results, clinical notes, diagnostic codes, and procedures. "
"Focus on determining whether the DRG assigned is accurate based on all available data and clinical evidence. "
"If discrepancies are found, provide a corrected DRG with an explanation. "
"Start your response by explaining what a DRG is and its significance in healthcare."
),
expected_output=(
"1) A brief explanation of what a DRG is and its significance in healthcare. "
"2) A concise validation of the DRG assigned to the patient, including: "
" a) A statement on whether the assigned DRG is correct. "
" b) If incorrect, the corrected DRG along with supporting evidence. "
" c) A brief explanation of the reasoning, citing clinical markers, symptoms, and procedures from the data."
"3) A summary of the key clinical indicators that influenced your decision."
"4) A final recommendation based on the analysis."
),
agent=identify_drg_gemini
)
# Define a task for Azure OpenAI
task_identify_drg_azure = Task(
description=(
f"Analyze the medical documents of the given patient to validate the Diagnosis-Related Group (DRG) assigned. Medical documents: {clinical_notes}. "
"Use tools to analyze the patient's clinical data, including lab results, clinical notes, diagnostic codes, and procedures. "
"Focus on determining whether the DRG assigned is accurate based on all available data and clinical evidence. "
"If discrepancies are found, provide a corrected DRG with an explanation. "
"Start your response by explaining what a DRG is and its significance in healthcare."
),
expected_output=(
"1) A brief explanation of what a DRG is and its significance in healthcare. "
"2) A concise validation of the DRG assigned to the patient, including: "
" a) A statement on whether the assigned DRG is correct. "
" b) If incorrect, the corrected DRG along with supporting evidence. "
" c) A brief explanation of the reasoning, citing clinical markers, symptoms, and procedures from the data."
"3) A summary of the key clinical indicators that influenced your decision."
"4) A final recommendation based on the analysis."
),
agent=identify_drg_azure
)
# Create a Crew instance
crew=Crew(
agents=[identify_drg_gemini, identify_drg_azure],
tasks=[task_identify_drg_gemini, task_identify_drg_azure],
process=Process.sequential,
)
# Execute the Crew instance
result=crew.kickoff(inputs={'topic':'Identify and validate the different DRGs for the patients based on comprehensive analysis.'})
Disclaimer: The posts here represent my personal views, not those of my employer or any specific vendor. Any technical advice or instructions are based on my knowledge and experience.