BigQuery and Postgres as Vector Databases
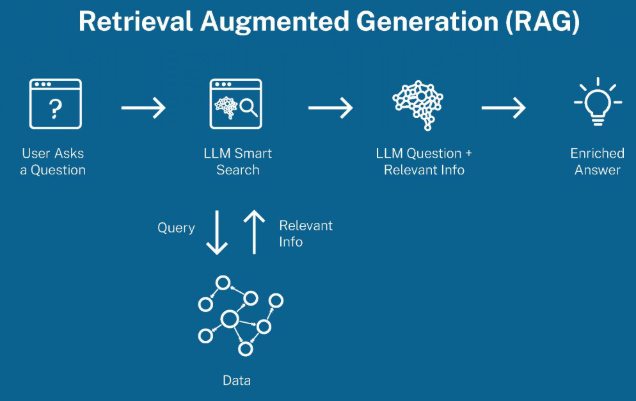
With the explosion of Large Language Models (LLMs) and their applications in Retrieval-Augmented Generation (RAG), the demand for efficient and scalable vector databases has never been greater. Unlike traditional databases that store and query structured data, vector databases are optimized for high-dimensional embeddings — numerical representations of text, images, and other unstructured data.
In RAG workflows, vector databases are the backbone for retrieving the most relevant context for an LLM, enabling more accurate and context-aware responses. Whether it’s enhancing chatbots, document search, or semantic search applications, vector search ensures that information retrieval is based on meaning rather than just keywords.
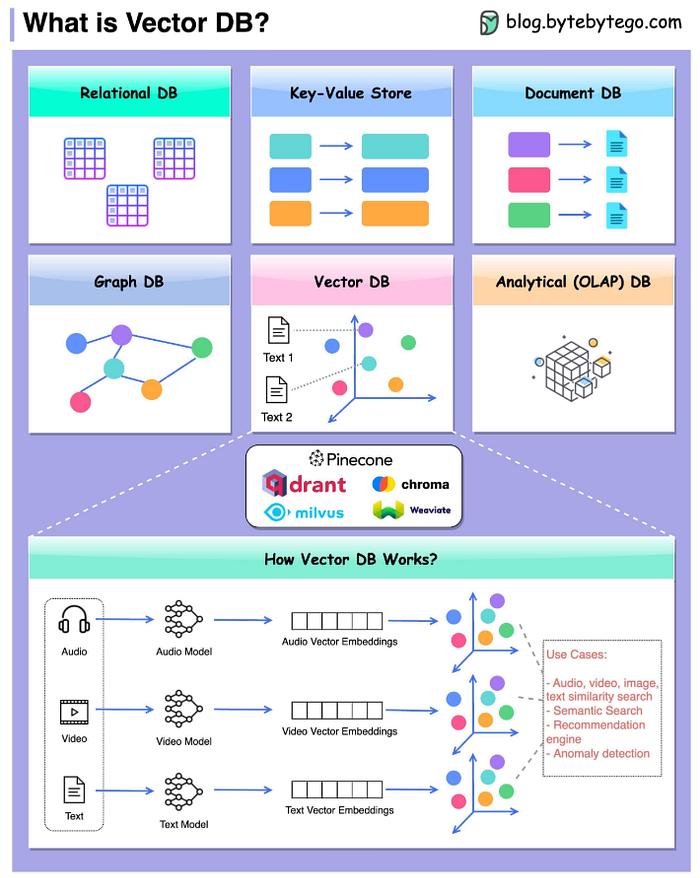
In this blog post, I share insights and code on utilizing GCP Cloud SQL for PostgreSQL and BigQuery as vector databases tailored to my specific use case.
BigQuery, Google Cloud’s serverless data warehouse, has recently launched vector search functionalities. This feature helps users efficiently store and query high-dimensional embeddings. Thanks to its scalability and compatibility with AI/ML tools, BigQuery supports rapid approximate nearest neighbor (ANN) searches, positioning it as an excellent option for RAG applications. It removes the requirement for database management, harnessing Google’s infrastructure for swift queries on vast datasets.
PostgreSQL is a popular open-source relational database that allows vector search via the pgvector extension. Google Cloud Platform’s Cloud SQL for PostgreSQL offers managed database services, including automatic scaling, backups, and robust security. This enables companies to store and query vector embeddings using exact or approximate search methods like IVFFlat and HNSW. Although it may not rival the speed of specialized vector databases, it balances flexibility, reliability, and the SQL interface familiar to PostgreSQL users.
Getting data from the MIMIC-III Clinical database
MIMIC-III is an extensive, freely available database comprising de-identified health-related data associated with over forty thousand patients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. More details about the database can be found here.
I ran this SQL statement to get the clinical notes of 100 patients. I exported the data to a CSV file and used it in my code, as PhysioNet doesn’t allow you to run BigQuery jobs directly on the MIMIC-III Clinical database.
WITH
patient_subset AS (
SELECT
DISTINCT subject_id
FROM
`physionet-data.mimiciii_clinical.patients`
ORDER BY
subject_id
LIMIT
100 ),
bnp_data AS (
SELECT
ce.subject_id,
ce.hadm_id,
ce.charttime AS bnp_time,
ce.valuenum AS bnp_level
FROM
`physionet-data.mimiciii_clinical.chartevents` ce
JOIN
`physionet-data.mimiciii_clinical.d_items` di
ON
ce.itemid = di.itemid
WHERE
LOWER(di.label) LIKE '%bnp%'
AND ce.valuenum IS NOT NULL ),
combined_notes AS (
SELECT
ne.subject_id,
ne.hadm_id,
STRING_AGG(ne.text, ' ') AS all_clinical_notes
FROM
`physionet-data.mimiciii_notes.noteevents` ne
JOIN
patient_subset ps
ON
ne.subject_id = ps.subject_id
GROUP BY
ne.subject_id,
ne.hadm_id )
SELECT
p.subject_id,
p.gender,
p.dob,
a.hadm_id,
a.admittime,
a.dischtime,
a.hospital_expire_flag,
dc.drg_code,
dc.description AS drg_description,
d.icd9_code AS diagnoses_icd9_codes,
bn.bnp_level,
bn.bnp_time,
cn.all_clinical_notes
FROM
patient_subset ps
JOIN
`physionet-data.mimiciii_clinical.patients` p
ON
ps.subject_id = p.subject_id
JOIN
`physionet-data.mimiciii_clinical.admissions` a
ON
p.subject_id = a.subject_id
LEFT JOIN
`physionet-data.mimiciii_clinical.drgcodes` dc
ON
a.hadm_id = dc.hadm_id
LEFT JOIN
`physionet-data.mimiciii_clinical.diagnoses_icd` d
ON
a.hadm_id = d.hadm_id
LEFT JOIN
bnp_data bn
ON
a.hadm_id = bn.hadm_id
LEFT JOIN
combined_notes cn
ON
a.hadm_id = cn.hadm_id;GCP Cloud SQL for PostgreSQL as vector database
# Using GCP Cloud SQL for PostgreSQL as Vector database via pgvector extension
### Import python packages
import google.auth
import numpy as np
import pandas as pd
import psycopg2
from sqlalchemy import create_engine
from vertexai.preview.language_models import TextEmbeddingModel
### GCP setup
_CREDENTIALS, _PROJECT_ID = google.auth.default()
print("Credentials:", _CREDENTIALS)
print("Project ID:", _PROJECT_ID)
_CREDENTIALS, _PROJECT_ID = google.auth.default()
_JSON_HEADERS = {"Content-Type": "application/fhir+json;charset=utf-8"}
_EMBED_MODEL = 'text-embedding-004'
### Reusable functions
# Function to generate text embeddings
def text_embedding(text: str) -> list[float]:
model = TextEmbeddingModel.from_pretrained(_EMBED_MODEL)
embeddings = model.get_embeddings([text])
return embeddings[0].values # Extract the embedding vector
def get_table_size(host, dbname, user, password):
"""
Returns the size of a PostgreSQL table in a human-readable format.
Parameters:
- table_name (str): Name of the table whose size you want to check.
- host (str): Hostname or IP address of the PostgreSQL instance.
- dbname (str): Name of the database.
- user (str): Username to connect to the PostgreSQL database.
- password (str): Password for the PostgreSQL user.
Returns:
- str: Human-readable size of the table.
"""
try:
# Create a cursor
cur = conn.cursor()
# SQL query to get table size
query = f"SELECT pg_size_pretty(pg_total_relation_size('mimic_drg')) AS table_size;"
# Execute the query
cur.execute(query)
# Fetch the result
table_size = cur.fetchone()
# Return the table size
return table_size[0]
except Exception as e:
return f"Error: {str(e)}"
finally:
# Close the cursor
if cur:
cur.close()
### Load the MIMIC-III data into GCP CloudSQL for PostgreSQL table
file_path = "/Users/Downloads/mimic_drg.csv"
clinical_notes = pd.read_csv(file_path, low_memory=False)
DB_USER = <database user name>
DB_PASSWORD = <database pwd>
DB_HOST = <database IP address>
DB_PORT = "5432"
DB_NAME = "mimic"
# Create a database connection
pg_engine = create_engine(f"postgresql://{DB_USER}:{DB_PASSWORD}@{DB_HOST}:{DB_PORT}/{DB_NAME}")
# # Verify the connection
df = pd.read_sql("SELECT 1", pg_engine)
print(df)
# Load DataFrames into PostgreSQL tables
clinical_notes.to_sql("mimic_drg", pg_engine, if_exists="replace", index=False)
print("Data successfully loaded into PostgreSQL!")
### Establish connection to PostgreSQL
conn = psycopg2.connect(
dbname="mimic",
user=<database user name>,
password=<database pwd>,
host=<database IP address>,
port="5432"
)
### Get the size of the mimic_drg table after the data load
table_size = get_table_size(DB_HOST, DB_NAME, DB_USER, DB_PASSWORD)
print(f"Table Size: {table_size}")
### Create and load embeddings into all_clinical_notes_embeddings column in the mimic_drg table
# Create the vector extension
cur = conn.cursor()
# Execute the SQL command
cur.execute("CREATE EXTENSION IF NOT EXISTS vector;")
conn.commit() # Commit changes
# Close the connection
cur.close()
print("Extension 'vector' enabled successfully.")
# Create the new column in mimic_drg table to store the embeddings
cur = conn.cursor()
# Execute the SQL command
cur.execute("ALTER TABLE mimic_drg ADD COLUMN IF NOT EXISTS all_clinical_notes_embeddings vector(768);")
conn.commit() # Commit changes
# Close the connection
cur.close()
print("New column is created successfully")
# Fetch all rows with clinical notes
cur = conn.cursor()
cur.execute("SELECT subject_id, hadm_id, all_clinical_notes FROM mimic_drg WHERE all_clinical_notes IS NOT NULL;")
rows = cur.fetchall()
# print("Fetched rows:", rows[:5]) # Print first few rows to debug structure
# Create and load embeddings
batch_size = 100 # Commit after every 100 rows
count = 0 # Track the number of processed rows
for row in rows:
try:
if len(row) != 3: # Ensure correct structure (subject_id, hadm_id, notes)
print(f"Skipping row due to unexpected structure: {row}")
continue
subject_id, hadm_id, notes = row
if not isinstance(notes, str) or not notes.strip():
print(f"Skipping subject_id {subject_id}: Invalid or empty notes")
continue
# Generate embedding
embedding_vector = text_embedding(notes)
embedding_str = "[" + ",".join(map(str, embedding_vector)) + "]"
# Update the mimic_drg table with the embedding
cur.execute("""
UPDATE mimic_drg
SET all_clinical_notes_embeddings = %s
WHERE subject_id = %s;
""", (embedding_str, subject_id))
count += 1
# Commit after every `batch_size` updates
if count % batch_size == 0:
conn.commit()
print(f"Committed {count} updates so far...")
except Exception as e:
print(f"Error processing subject_id {subject_id}: {e}")
# Final commit for any remaining updates
if count % batch_size != 0:
conn.commit()
cur.close()
print("All embeddings processed and updated successfully!")
# Size of the table after adding the embeddings
table_size = get_table_size(DB_HOST, DB_NAME, DB_USER, DB_PASSWORD)
print(f"Table Size: {table_size}")
### Create an embedding for the query used in the SELECT statements
# Use the generated query vector for 'pulmonary edema'
query_vector = text_embedding("pulmonary edema")
# Format the query vector as a string (replace this with the actual vector you get)
query_vector_str = str(query_vector).replace('[', '').replace(']', '')
print("Query Vector:", query_vector_str)
### Create IVVFLAT index on all_clinical_notes_embeddings column in mimic_drg table with different Distance Functions
def create_index(conn, cur, index_name, create_index_sql):
cur = conn.cursor()
cur.execute(create_index_sql)
conn.commit()
cur.close()
# Define SQL statements for each index creation
index_creation_sqls = [
{
'name': 'mimic_drg_embeddings_ivfflat_l2',
'sql': """
CREATE INDEX IF NOT EXISTS mimic_drg_embeddings_ivfflat_l2
ON mimic_drg USING ivfflat (all_clinical_notes_embeddings vector_l2_ops)
WITH (lists = 100);
"""
},
{
'name': 'mimic_drg_embeddings_ivfflat_ip',
'sql': """
CREATE INDEX IF NOT EXISTS mimic_drg_embeddings_ivfflat_ip
ON mimic_drg USING ivfflat (all_clinical_notes_embeddings vector_ip_ops)
WITH (lists = 100);
"""
},
{
'name': 'mimic_drg_embeddings_ivfflat_cosine',
'sql': """
CREATE INDEX IF NOT EXISTS mimic_drg_embeddings_ivfflat_cosine
ON mimic_drg USING ivfflat (all_clinical_notes_embeddings vector_cosine_ops)
WITH (lists = 100);
"""
}
]
# Establish connection and cursor (assuming conn and cur are already defined)
for index in index_creation_sqls:
create_index(conn, cur, index['name'], index['sql'])
# Size of the table after adding the indexes
table_size = get_table_size(DB_HOST, DB_NAME, DB_USER, DB_PASSWORD)
print(f"Table Size: {table_size}")
### Run SQL statements on mimic_drg table with IVVFLAT indexes
cur = conn.cursor()
# SQL query to search for the most similar notes based on the embeddings usiing the Cosine distance operator
sql_query = f"""
SELECT subject_id, hadm_id, gender, dob, admittime, dischtime, drg_code, drg_description, diagnoses_icd9_codes, all_clinical_notes
FROM mimic_drg
ORDER BY all_clinical_notes_embeddings <=> '{query_vector}'
limit 5;
"""
# Execute the query
cur.execute(sql_query)
results = cur.fetchall()
cur.close()
# Print results
for result in results:
print(result)
cur = conn.cursor()
# SQL query to search for the most similar notes based on the embeddings using the Euclidean (L2 Distance) distance operator
sql_query = f"""
SELECT subject_id, hadm_id, gender, dob, admittime, dischtime, drg_code, drg_description, diagnoses_icd9_codes, all_clinical_notes
FROM mimic_drg
ORDER BY all_clinical_notes_embeddings <-> '{query_vector}'
limit 5;
"""
# Execute the query
cur.execute(sql_query)
results = cur.fetchall()
cur.close()
# Print results
for result in results:
print(result)
cur = conn.cursor()
# SQL query to search for the most similar notes based on the embeddings using the Inner Product distance operator
sql_query = f"""
SELECT subject_id, hadm_id, gender, dob, admittime, dischtime, drg_code, drg_description, diagnoses_icd9_codes, all_clinical_notes
FROM mimic_drg
ORDER BY all_clinical_notes_embeddings <#> '{query_vector}'
limit 5;
"""
# Execute the query
cur.execute(sql_query)
results = cur.fetchall()
cur.close()
# Print results
for result in results:
print(result)
### Drop IVVFLAT indexes
**It's not a best practice to force the query optimizer to choose a particular index. So, I'm dropping IVVFLAT indexes to test HNSW indexes.**
cur = conn.cursor()
# Execute the SQL command
cur.execute("DROP INDEX mimic_drg_embeddings_ivfflat_l2;")
conn.commit() # Commit changes
cur.execute("DROP INDEX mimic_drg_embeddings_ivfflat_ip;")
conn.commit() # Commit changes
cur.execute("DROP INDEX mimic_drg_embeddings_ivfflat_cosine;")
conn.commit() # Commit changes
# Close the connection
cur.close()
### Create HNSW index on all_clinical_notes_embeddings column in mimic_drg table with different Distance Functions
def create_index(query, description):
cur = conn.cursor()
cur.execute(query)
conn.commit()
cur.close()
# Define SQL queries for each index
hnsw_l2_query = """
CREATE INDEX mimic_drg_embeddings_hnsw_l2
ON mimic_drg USING hnsw (all_clinical_notes_embeddings vector_l2_ops);
"""
hnsw_ip_query = """
CREATE INDEX mimic_drg_embeddings_hnsw_ip
ON mimic_drg USING hnsw (all_clinical_notes_embeddings vector_ip_ops);
"""
hnsw_cosine_query = """
CREATE INDEX mimic_drg_embeddings_hnsw_cosine
ON mimic_drg USING hnsw (all_clinical_notes_embeddings vector_cosine_ops);
"""
create_index(hnsw_l2_query, "HNSW L2 Distance Index Creation")
create_index(hnsw_ip_query, "HNSW Inner Product Index Creation")
create_index(hnsw_cosine_query, "HNSW Cosine Distance Index Creation")
### Run SQL statements on mimic_drg table with HNSW indexes
cur = conn.cursor()
# SQL query to search for the most similar notes based on the embeddings usiing the Cosine distance operator
sql_query = f"""
SELECT subject_id, hadm_id, gender, dob, admittime, dischtime, drg_code, drg_description, diagnoses_icd9_codes, all_clinical_notes
FROM mimic_drg
ORDER BY all_clinical_notes_embeddings <=> '{query_vector}'
limit 5;
"""
# Execute the query
cur.execute(sql_query)
results = cur.fetchall()
cur.close()
# Print results
for result in results:
print(result)
cur = conn.cursor()
# SQL query to search for the most similar notes based on the embeddings using the Euclidean (L2 Distance) distance operator
sql_query = f"""
SELECT subject_id, hadm_id, gender, dob, admittime, dischtime, drg_code, drg_description, diagnoses_icd9_codes, all_clinical_notes
FROM mimic_drg
ORDER BY all_clinical_notes_embeddings <-> '{query_vector}'
limit 5;
"""
# Execute the query
cur.execute(sql_query)
results = cur.fetchall()
cur.close()
# Print results
for result in results:
print(result)
cur = conn.cursor()
# SQL query to search for the most similar notes based on the embeddings using the Inner Product distance operator
sql_query = f"""
SELECT subject_id, hadm_id, gender, dob, admittime, dischtime, drg_code, drg_description, diagnoses_icd9_codes, all_clinical_notes
FROM mimic_drg
ORDER BY all_clinical_notes_embeddings <#> '{query_vector}'
limit 5;
"""
# Execute the query
cur.execute(sql_query)
results = cur.fetchall()
cur.close()
# Print results
for result in results:
print(result)
### Close the Postgres connection
conn.close()BigQuery as vector database
# Using GCP BigQuery as Vector database
### Import python packages
import google.auth
from google.cloud import bigquery
import numpy as np
import pandas as pd
from vertexai.preview.language_models import TextEmbeddingModel
### GCP setup
_CREDENTIALS, _PROJECT_ID = google.auth.default()
print("Credentials:", _CREDENTIALS)
print("Project ID:", _PROJECT_ID)
_CREDENTIALS, _PROJECT_ID = google.auth.default()
_JSON_HEADERS = {"Content-Type": "application/fhir+json;charset=utf-8"}
_EMBED_MODEL = 'text-embedding-004'
### Reusable functions
# Function to generate text embeddings
def text_embedding(text: str) -> list[float]:
model = TextEmbeddingModel.from_pretrained(_EMBED_MODEL)
embeddings = model.get_embeddings([text])
return embeddings[0].values # Extract the embedding vector
def get_table_size():
"""
Returns the size of a BigQuery table in a human-readable format.
Parameters:
- project_id (str): GCP Project ID.
- dataset_id (str): BigQuery Dataset ID.
- table_id (str): BigQuery Table ID.
Returns:
- str: Human-readable size of the table.
"""
try:
client = bigquery.Client(project=project_id)
# Fetch table metadata
table_ref = f"<project_id>.mimic_drg.mimic_drg_clinical_notes"
table = client.get_table(table_ref)
# Get table size in bytes and convert to human-readable format
table_size_bytes = table.num_bytes
table_size_mb = table_size_bytes / (1024 * 1024) # Convert to MB
return f"{table_size_mb:.2f} MB"
except Exception as e:
return f"Error: {str(e)}"
### Load the MIMIC-III data into GCP BigQuery dataset
file_path = "/Users/Downloads/mimic_drg.csv"
clinical_notes = pd.read_csv(file_path, low_memory=False)
# Define parameters
project_id = <project_id>
dataset_id = "mimic_drg"
table_id = "mimic_drg_clinical_notes"
# Define the full table reference
table_ref = f"{project_id}.{dataset_id}.{table_id}"
from google.cloud.bigquery import LoadJobConfig, WriteDisposition
# Initialize BigQuery client
client = bigquery.Client(project=project_id)
# Load DataFrame into BigQuery
job_config = LoadJobConfig(write_disposition=WriteDisposition.WRITE_TRUNCATE)
job = client.load_table_from_dataframe(clinical_notes, table_ref, job_config=job_config)
# Wait for the job to complete
job.result()
# Print success message
print(f"CSV file successfully loaded into BigQuery table: {table_ref}")
### Get the size of the mimic_drg_clinical_notes table after the data load
table_size = get_table_size()
print(f"Table Size: {table_size}")
### Create embeddings for all_clinical_notes_embeddings column in the mimic_drg_clinical_notes table
from google.cloud import bigquery_connection_v1 as bq_connection
def create_external_connection(project_id, connection_id):
"""Creates an external connection in BigQuery."""
connection_client = bq_connection.ConnectionServiceClient()
parent = f"projects/{project_id}/locations/us"
connection = bq_connection.Connection()
connection.cloud_resource = bq_connection.CloudResourceProperties()
connection.name = connection_client.connection_path(project_id, "us", connection_id)
request = bq_connection.CreateConnectionRequest(
parent=parent, connection_id=connection_id, connection=connection
)
try:
response = connection_client.create_connection(request=request)
print(f"External connection '{connection_id}' created successfully: {response.name}")
return True
except Exception as e:
print(f"Error creating external connection: {e}")
return False
create_external_connection(project_id, connection_id)
# Initialize BigQuery client
client = bigquery.Client(project=project_id)
# Construct the fully qualified connection name
connection_name = f"projects/{project_id}/locations/us/connections/mimic_drg" # Replace "us" with your location if needed
# Define the SQL statement with the fully qualified connection name
query = f"""
CREATE OR REPLACE MODEL `{project_id}.{dataset_id}.mimic_embedding_model`
REMOTE WITH CONNECTION `{connection_name}`
OPTIONS (ENDPOINT = 'text-embedding-004');
"""
# Run the query
query_job = client.query(query)
query_job.result() # Wait for the query to complete
print("Remote model created successfully.")
# Initialize BigQuery client
client = bigquery.Client(project=project_id)
# Step 1: Generate embeddings and store in a temporary table
temp_table = f"{project_id}.{dataset_id}.mimic_drg_temp_embeddings"
query_generate_embeddings = f"""
CREATE OR REPLACE TABLE `{temp_table}` AS
(SELECT * FROM
ML.GENERATE_TEXT_EMBEDDING(
MODEL `{project_id}.{dataset_id}.mimic_embedding_model`,
(SELECT *, all_clinical_notes AS content FROM `{table_ref}`)
)
);
"""
# Run the query to generate embeddings
query_job = client.query(query_generate_embeddings)
query_job.result() # Wait for the query to complete
print("Embeddings generated and stored in temporary table.")
### Create an embedding for the query used in the SELECT statements
# Use the generated query vector for 'pulmonary edema'
query_vector = text_embedding("pulmonary edema")
# Format the query vector as a string (replace this with the actual vector you get)
query_vector_str = str(query_vector).replace('[', '').replace(']', '')
print("Query Vector:", query_vector_str)
### Run SQL statements on mimic_drg_temp_embeddings table
def vector_search():
"""
Performs a vector search using the specified model.
Args:
project_id: Your Google Cloud project ID.
dataset_id: The ID of your BigQuery dataset.
"""
client = bigquery.Client(project=project_id)
query = f"""
SELECT
base.subject_id,
base.hadm_id,
base.gender,
base.dob,
base.admittime,
base.dischtime,
base.drg_code,
base.drg_description,
base.diagnoses_icd9_codes,
base.all_clinical_notes,
distance
FROM
VECTOR_SEARCH(
TABLE `{project_id}.{dataset_id}.mimic_drg_temp_embeddings`,
'text_embedding',
(SELECT ARRAY<FLOAT64>{query_vector} AS query_embedding),
distance_type => 'cosine'
);
"""
try:
query_job = client.query(query)
results = query_job.result()
for row in results:
print(row.subject_id, row.hadm_id, row.gender,row.dob, row.admittime, row.dischtime, row.drg_code, row.drg_description, row.diagnoses_icd9_codes,row.all_clinical_notes, row.distance)
except Exception as e:
print(f"Error executing query: {e}")
vector_search()Disclaimer: The posts here represent my personal views, not those of my employer or any specific vendor. Any technical advice or instructions are based on my knowledge and experience.